Tag: Bacteria
-
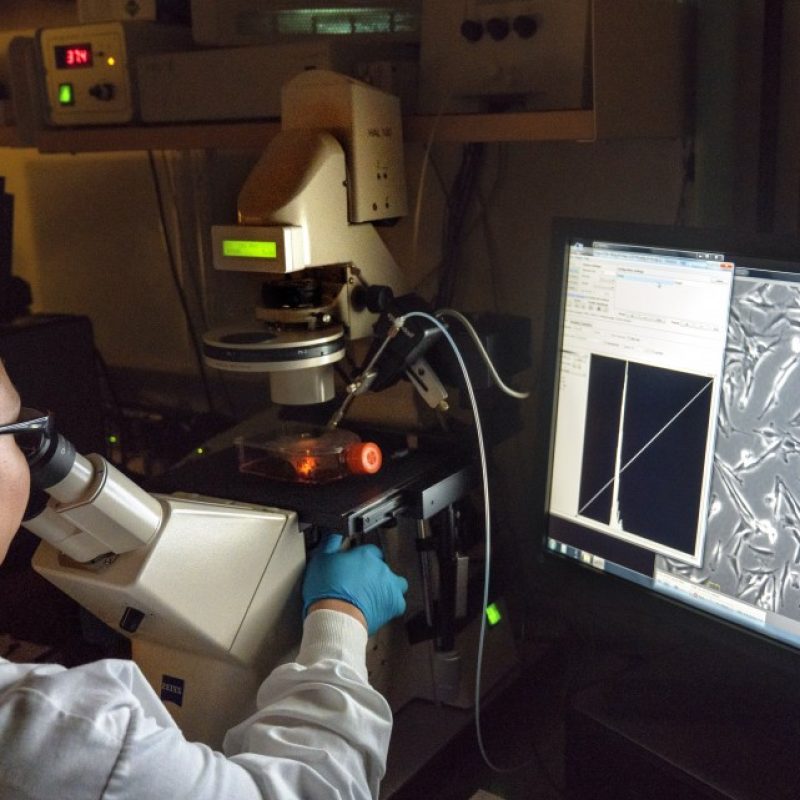
Whittaker lab: Coronaviruses and Musteloidea
The Whittaker lab’s paper out in mBio that looks deeper into coronaviruses and their relationship to the superfamily Musteloidea. PAPER: https://mbio.asm.org/content/12/1/e02873-20
-
Pawlowska: Food safety and fungi
Assoc. Prof. Teresa Pawlowska discusses her lab’s work on food safety and fungi. The full text of the Cornell Chronicle story is below. CORNELL CHRONICLE STORY By Krishan Ramanujan A new grant will investigate how bacteria that live inside the cells of fungi may shape the biology, evolution, biodiversity and function of these fungi – […]
-

Corrie Moreau: Sharing 7 million insects with the digital world
Prof. Corrie Moreau talks about digitizing the Cornell University Insect Collection. The full article appears below. CALS ARTICLE By Krishna Ramanujan As director and head curator of the Cornell University Insect Collection, Corrie Moreau has numerous tasks on her to-do list, including one that could last her entire career: digitizing the collection’s 7 million specimens. Digitization is a […]
-

Helmann lab: new elongasome function paper
The John Helmann lab has a new paper out, “A regulatory pathway that selectively up-regulates elongasome function in the absence of class A PBPs” in eLife. LINK TO PAPER: https://pubmed.ncbi.nlm.nih.gov/32897856/
-

Evolution of Symbiosis
The long-term goals served by this project are mechanistic understanding of plant disease and development of broadly effective and durable means of control. The project seeks to structurally and functionally characterize a pathogen-activated host gene that plays a critical role in disease in a major crop species, and to ascertain the potential of strategies to […]
-

Pamela Chang Lab
The Chang lab conducts research at the interface of chemical biology, microbiology, and immunology. Our research is focused on understanding chemical communication between the gut microbiome and the host immune system. Using both chemical and biological approaches, we develop novel chemical tools to understand 1) the metabolism of the gut microbiome and 2) important pathways […]
-

Molecular dialogue between intestinal stem cells and microbiota
The intestinal epithelium faces unique challenges as it is constantly exposed to the passage of ingested material including food, bacteria and xenobiotics. To maintain tissue function, the intestinal epithelium is undergoing continuous renewal mediated by intestinal stem cells (ISCs). ISC proliferation and differentiation are constantly adapted both to the microbes present and to the gut […]
-

Development of broadly neutralizing antibodies to influenza
Development of broadly neutralizing antibodies to influenza using a novel bacterial outer membrane vesicle platform: The Leifer and Putnam labs have an ongoing collaboration to use innovative engineering techniques develop, test, and understand the underlying immunological mechanisms of new vaccine adjuvant bacterial outer membrane vesicle platforms. We are interested in postdoctoral candidates proposing to take […]
-

Ecology and Evolution in Bacteria-Host Interactions
The Hendry lab uses experimental and -omics approaches to understand the impact of host interactions on bacterial ecology and evolution, as well as how microbes influence hosts. Research in the lab focuses on a variety of systems, particularly insect and plant associated bacteria. Our interdisciplinary group encompasses broad interests within microbiology, evolution, and ecology and […]
-

Fungal-bacterial interactions
The Pawlowska Lab studies interactions between fungi and bacteria using several model systems. Potential projects involve: (1) dissecting the mechanisms of innate immunity in fungi, (2) evaluating the impact of bacteriome on phenotypic diversity of fungi, and (3) examining the role of common mycorrhizal networks in plant microbiome assembly.
